Please note that this is the legacy page of the 2nd Challenge, which concluded in Feb 2024.
Welcome to the CMI-PB Challenge, a systems immunology competition. This is an exciting opportunity to explore the world of systems vaccinology by analyzing longitudinal immune response data obtained through cutting-edge multi-omics experiments. We have established a resource based on our Computational Models of Immunity to Pertussis Booster vaccinations (CMI-PB) Project to generate experimental data to create and test computational models that predict vaccination outcomes based on the baseline state of the vaccines.
Overview
The overall goal of the CMI-PB prediction challenge is two-fold. We want to:
- Establish a community platform to test and compare computational models of immunity in vaccination.
- Better understand vaccine-induced immunity to B. pertussis. By establishing and testing computational models that attempt to predict the cascade of events that follow B. pertussis booster vaccination, we will improve our understanding of the mechanisms underlying these events, with the ultimate goal of identifying what variables induce a strong and durable recall response.
The CMI-PB prediction challenge is an exciting opportunity to explore the complex world of systems vaccinology by utilizing longitudinal immune response data obtained through cutting-edge multi-omics experiments. With this challenge, the CMI-PB prediction contest hopes to foster a collaborative research community, addressing challenges and advancing scientific knowledge more rapidly than any individual or research group could achieve alone. We have successfully concluded the 1st challenge with contestants from the CMI-PB member labs and published the results here. For the second challenge, we are expanding the participant pool by inviting specific contestants outside the CMI-PB network before rolling out the public challenge.
Your task during this challenge is to predict outcomes, showcasing your intuition and analytical skills. To participate in the second CMI-PB prediction challenge, utilize the provided training dataset to build your computational models and predict the vaccination outcomes of newly tested individuals (test dataset). To create a submission, use predictive computational modeling techniques to provide answers to the list of prediction tasks. The CMI-PB team will evaluate submissions from all the contestants and inform you of the outcome of the contest. We recommend checking out the 'Learn about the project' section to delve deeper into the topic. We have also provided teaching materials to explain background and theme of the project.
When you're ready to begin, create an account and click the 'Submit Prediction' button in the upper right-hand corner of this page. To find these buttons, you must first scroll back up to the top of this page. No login is necessary to access the competition data. However, login is required to record and track your submissions. We look forward to your participation!

Figure 1: CMI-PB Challenge Outline
Please see the CMI-PB Prediction Challenge timeline below.
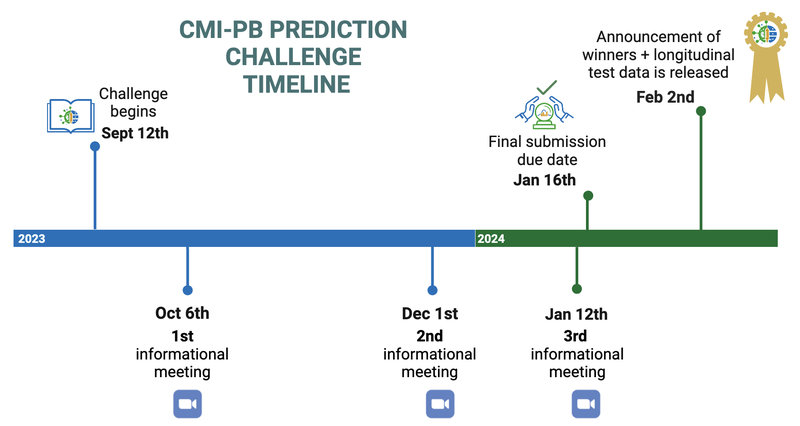
Figure 2: Prediction Challenge timeline
Data and resources
Study design and multi-omics datasets
Our cohort comprises of acellular Pertussis (aP) vs whole cell Pertussis (wP) infancy-primed subjects boosted with Tdap. We recruited individuals born before 1995 (wP) and after 1996 (aP), collected baseline plasma and blood samples, provided the Tdap booster vaccine, and then obtained plasma/blood at 1, 3, 7, and 14 days post booster vaccination. We recommend checking out the 'Learn about the project' and 'Understand the data' sections to delve deeper into study design, experimental data generation and standardization.
From the samples, we generated omics data by:
- Cell frequency in PBMCs (30+ cell populations) using flow cytometry,
- PBMC Gene expression (50,000+ genes),
- Plasma cytokine concentrations (30 soluble proteins) using Olink, which provides a quantitative readout of cytokines, chemokines, and other immune factors,
- Plasma Ab titers (7+ antigens)
Challenge data
The data has been split into two groups:
- Train/Model building set (Baseline and longitudinal readouts: 2020, 2021 dataset). To build your computational models, use the training set, which includes the outcome (also known as the "ground truth") for each subject. Your model will be based on features extracted from longitudinal multi-omics readouts (such as transcriptomics, proteomics, antibody titers, and cell frequency) as well as demographic data (such as age, infancy vaccination and biological sex). You can create new features using feature engineering techniques.
- Test/Prediction set (Baseline readouts: 2022 dataset). To evaluate your model's performance on new, unseen baseline data, use the test set. The test set does not provide the ground truth (longitudinal vaccine response) for each subject. Your task is to predict these outcomes using the model you built. Use your model to predict the vaccine response for each subject in the test set.
Data files download:
Raw Datasets are available via API and HTTPS file servers:
Raw datasets can be downloaded using API endpoints or by directly downloading the files (sftp site) from the following locations.
- Datasets: Training datasets | Prediction dataset
The CMI-PB team conducted data harmonization and processing to provide datasets in the form of computable matrices. These can be directly utilized for model building.
- Resources:
- Processed training datasets (R objects and TSV files) | Codebase: [Rpubs] [GitHub]
- Processed prediction datasets (R objects and TSV files) | Codebase: [Rpubs] [GitHub]
Important Note: As the 2nd challenge progresses, contestants might notice inconsistencies or issues in the dataset. It’s natural for challenge datasets to undergo modifications over time. We provided a dedicated page that organizes and tracks all changes related to the datasets. The tracking page is available here. Ensure you utilize the latest dataset when constructing your models.
Prediction challenge tasks
Overview:
Pre-vaccination information serves as crucial baseline data for predicting vaccine response. This includes:
1) Demographic data
- Age
- Biological sex at birth
- Vaccine priming status
2) Assays characterizing the immune status just before (baseline) and after the vaccine is given:
- Antibody titers in plasma by Luminex
- Secreted cytokine levels in plasma by OLINK
- Gene expression in PBMC by RNA-Seq
- Cell type frequencies in PBMC by Cytometry
List of Prediction challenge tasks:
1) Antibody titer tasks
1.1) Rank the individuals by IgG antibody titers against pertussis toxin (PT) that we detect in plasma 14 days post booster vaccinations.
1.2) Rank the individuals by fold change of IgG antibody titers against pertussis toxin (PT) that we detect in plasma 14 days post booster vaccinations compared to titer values at day 0.
2) Cell frequencies tasks
2.1) Rank the individuals by predicted frequency of Monocytes on day 1 post boost after vaccination.
2.2) Rank the individuals by fold change of predicted frequency of Monocytes on day 1 post booster vaccination compared to cell frequency values at day 0.
3) Gene expression tasks
3.1) Rank the individuals by predicted gene expression of CCL3 on day 3 post-booster vaccination.
3.2) Rank the individuals by fold change of predicted gene expression of CCL3 on day 3 post booster vaccination compared to gene expression values at day 0.
Examples of models
- We generated 32 computational models during 1st (internal) challenge. These models can be accessible here
- We also generated sample models on the 2nd challenge training dataset for demonstration purposes. These models can be accessible here
- Demonstration of model construction and submission file preparation process here
Submission instructions
Creating an account
- You must login/register to enter a submission into a challenge. After logging in, you can download the data and make a submission using the steps outlined below.
- An "Entry" is complete and will be evaluated when the data is submitted in the layout and tsv format specified on the website on the prediction task page.
- All entries must be during the competition period, displayed on the prediction task page.
- The ultimate goal is to model as many of the tasks as possible and submit your prediction by the due date.

Figure 3: Creating an account
Submission restrictions
- You are allowed 1 final entry, per account, but can re-submit multiple times (Max: 5 submissions) until the deadline. Note that your last submission will be considered your ‘final’ version (final entry).
- If you have developed multiple modeling approaches and wish to enter multiple entries, please create separate CMI-PB login accounts to manage these entries.
- For those utilizing more than three modeling strategies, please reach out to the CMI-PB team via email at cmi-pb-challenge@lji.org if there are any issues.
- External data is not allowed. Participants agree to make no attempt to use additional data or data sources not provided.
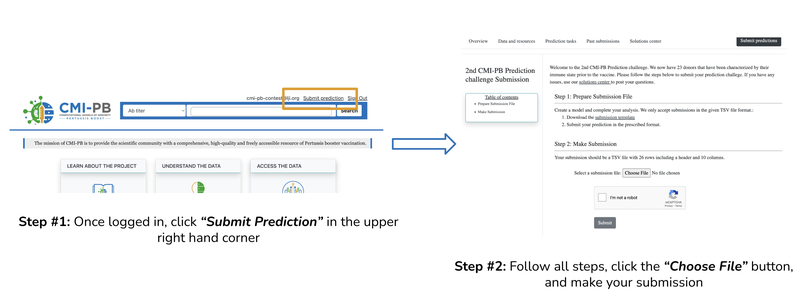
Figure 4: Submission process
Preparing a submission file
A complete submission file contains a total of 10 columns, consisting of 4 metadata columns and 6 columns dedicated to prediction tasks, with each column corresponding to a specific prediction task. The ultimate goal is to model as many of the tasks as possible, however contestants doesn't require to submit answers for all tasks. We expect contestants to generate computational models and upon making predictions these values are ranked from highest to lowest (i.e. highest = 1, lowest = N) before making a final submission.
- Submissions: Template file | Sample/demo submissions
In instances where the submission file lacks predictions for specific subjects, we will employ the median rank, which is calculated from the ranked list submitted by the contestant, to fill in the missing ranks.
Solutions Center
For Competition Help: CMI-PB solutions center
CMI-PB has a dedicated support team, so you’ll typically find that you receive a response more quickly by asking your question in the solutions center. The forum will work as a resource full of useful information on the data, metrics, and different approaches. We encourage you to use the forums often in an effort to increase collaboration.
Winner Announcement
The 2nd prediction challenge has successfully concluded on January 16, 2024. We received 26 outstanding submissions, and after careful consideration and evaluation, we are pleased to announce the winner for the 2nd CMI-PB challenge.
- 2nd CMI-PB Challenge Winner: Saonli Basu, Michael Anderson, Josey Sorenson, Katherine Li, Bhargob Kakoty, and Cheng-Chang Wu, University of Minnesota Team (submission ID: user49)
We also wanted to acknowledge Joe Hou (Submission ID: user47) with the Curiosity Conductor Prize for utilizing the Solutions Center and helping to advance the CMI-PB Challenge. We appreciate your engagement and involvement in asking questions that helped the other participants.
We thank all participants for being a part of the 2nd CMI-PB Challenge. Your contributions and participation have made this contest a success, and we look forward to your continued involvement in CMI-PB public challenges. All models submitted during 2nd CMI-PB challenge were made publically accessible via Github https://github.com/topics/cmipb-challenge. We are currently drafting a manuscript summarising all models submitted during the 2nd challenge. We will share more information about it soon.